一种基于Droplet microfluidics的ELISA方法。
Introduction
蛋白质是一种重要的生物标记物,很多蛋白质在生物样本中含量很低,因此需要超灵敏的蛋白检测技术。数字酶联免疫吸附试验(enzyme-linked immunosorbent assay, ELISA)比传统ELISA灵敏度提高了1000倍,但是仍然在很多临床应用中有失灵敏。因此需要开发更灵敏的方法。
在使用单分子阵列(Simoa)的数字ELISA中,使用了远超于靶分子的抗体包被的小珠,因此大多数小珠不会结合任何靶分子。单个蛋白质分子被捕获在小珠上。然后用酶标记蛋白质,然后将小珠分离到飞升大小的孔中,使每个孔只能容纳一个小珠。用油密封阵列后,含有酶标记免疫复合物的孔会产生局部高浓度的荧光产物,从而可以通过对活性孔进行计数来检测单分子。为了确保最大的灵敏度,有必要分析许多小珠,然后分析阳性事件。通常使用外力来促进小珠装入孔中。在Simoa HD-1分析仪中,小珠是通过重力在孔内隔离的。然而,只有约5%的小珠被分离,导致灵敏度下降。可以通过施加磁力来提高顺磁性小珠的装载效率,也有人则使用了电场并显示了珠粒装载量的改善。但是,对免疫复合物形成和测定灵敏度提高的影响尚未得到广泛研究,可以提高珠粒装载效率且灵敏度提高的简单方法仍然需要开发。
油包水液滴的优点如下:
- 能以最小的小珠损失捕获高百分比的小珠进行分析,提高灵敏度。
- 液滴产生装置简单,不需要昂贵而复杂的精细结构。
- 能用一个简单的光学设置对液滴成像。
- 液滴产生很快,在几分钟内能轻易地产生数百万液滴。
他们的液滴数字ELISA(ddELISA)方法灵敏度相比于Simoa提高了25倍,达阿摩尔级(aM或10-18M)。
Figure 1. Single molecule protein detection using ddELISA
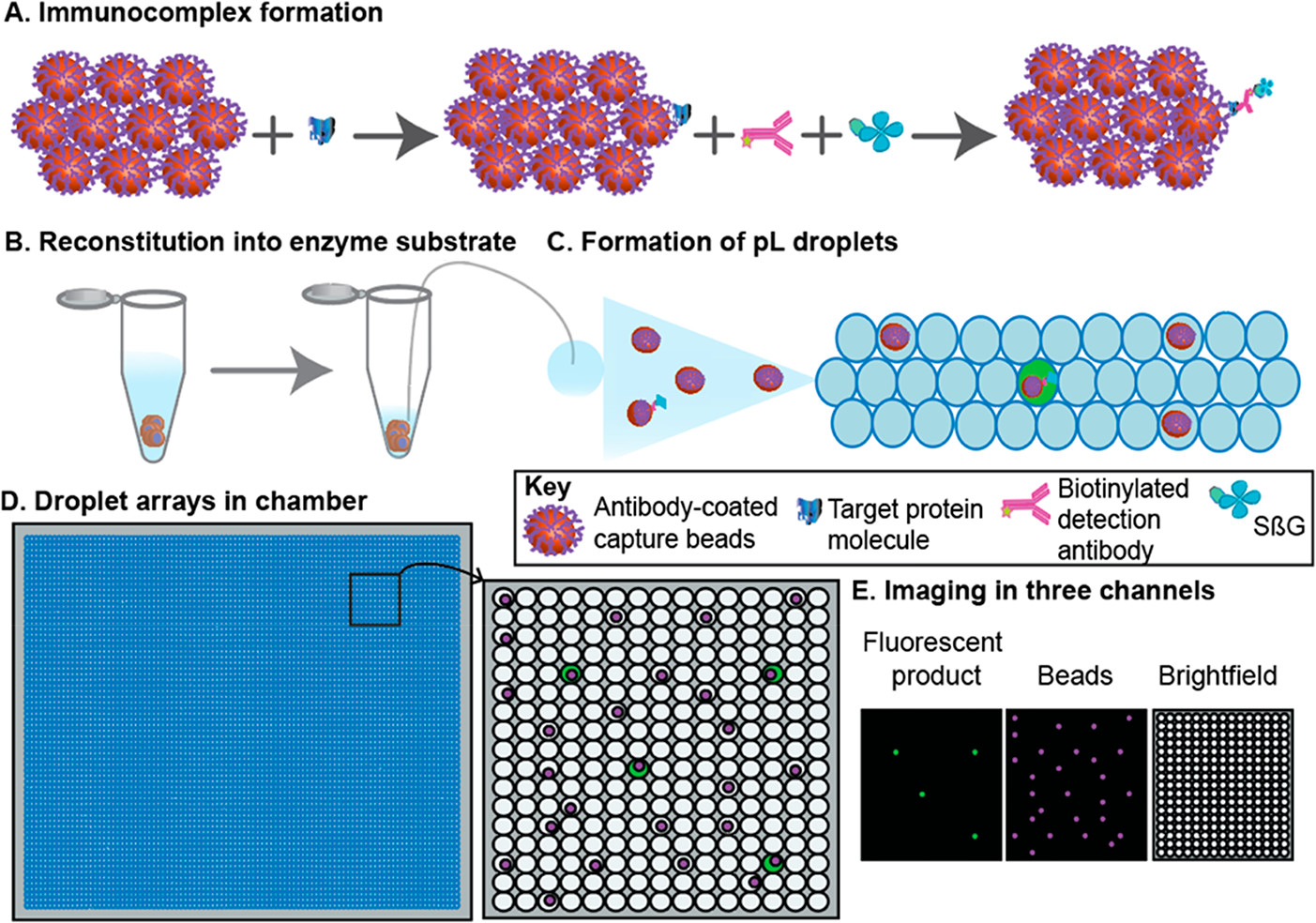
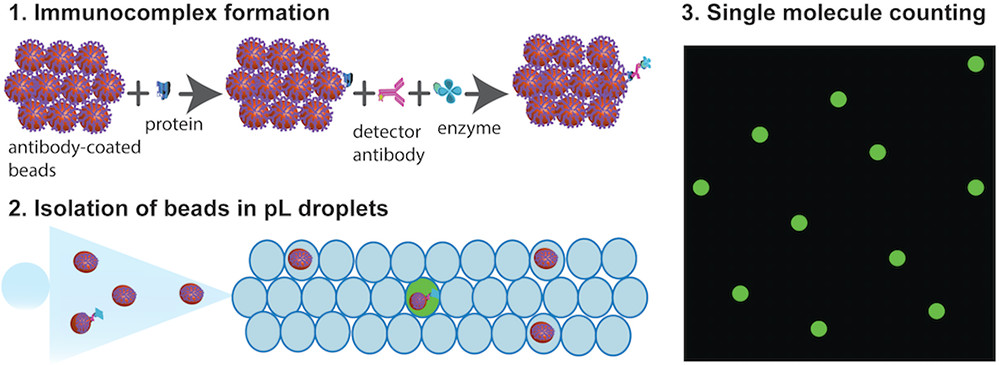
- (A)将100,000个抗体包被的顺磁珠添加到包含靶蛋白分子的样品中。然后加入生物素化的检测抗体和SβG,并与靶蛋白分子结合,形成酶标记的免疫复合物。根据泊松统计,大多数磁珠将具有零个免疫复合物,而一小部分将具有一个免疫复合物。
- (B)然后将小珠重新溶解在2μL的酶底物中。
- (C)然后将包含小珠和底物的2μL混合物划分到皮升级液滴中。
- (D)将液滴装载到腔室中,形成液滴阵列。
- (E)在三个通道中获得图像,以识别“含靶蛋白”的液滴,小珠和液滴。
双抗体夹心法,是将含有已知抗体的抗血清吸附在微量滴定板上的小孔里,洗涤一次;加待测抗原,如两者是特异的,则发生结合,然后把多余抗原洗除;加入与待测抗原呈特异反应的酶联抗体,使一单位的抗原同时结合两单位的抗体形成“夹心”;最后加入该酶的底物,根据产生的有色酶解产物颜色的深浅来判断待测抗原的含量。
作者认为的优势:
- 小珠的数量比Simoa的500,000减少到了100,000,从而有更高的fon ,信号也更高。此外,更方便即时应用。传统的微孔阵列则很难如此减少,因为对可分析小珠的数量有要求。
- 数字读数系统,小珠被包裹于皮升级的液滴中而不是飞升级的小孔。
- 使用的FDG底物比Simoa的RGP在液滴里更稳定。
Figure 2. Theoretical calculations

- (A)指定浓度下,小珠数目增多,每珠平均酶(AEB) 减少。
- (B)指定浓度下,分析的小珠百分比的越高,阳性事件数越高。
- (C)有限检测极限(LOD)随分析小珠分数而降低,即敏感度越高。此外,离解常数(KD)越低,敏感度越高。
Figure 3. ddELISA device design

- (A)1为油和表面活性剂入口,2为小珠和底物混合物入口,3为液珠出口。通道连接处宽度为9 μm,连接处最小特征为5 μm。整个装置尺寸为4.5 mm × 7 mm。
- (B)蓝框显示了阻塞柱,用于防止液滴(直径14 μm)逸出。两个立柱6之间的距离为7 μm。绿框显示了132个子室之一。柱4直径为60 μm,柱5之间为20 μm。整个腔室的尺寸为16 mm × 24 mm。
表面活性剂起阻止液滴之间融合的作用。
Figure 4. Calibration curves

- (A)IFNγ和IL-2的ddELISA校准曲线。
- (B)IFNγ和IL-2的Simoa校准曲线。
- (C)两种方法IFNγ和IL-2相对于背景的信号强度比较,底部为局部放大。可见ddELISA有更好的信号增加。
Figure 5. Endogenous protein measurements in serum
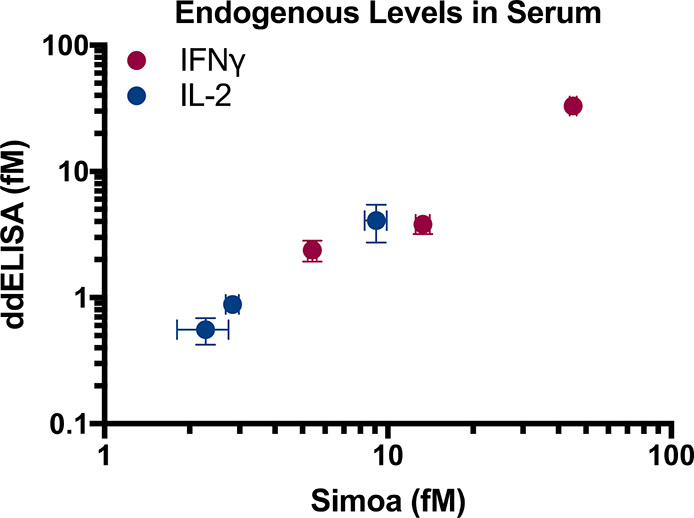
分别用两种方法测试三个样本每Marker,Pearson相关系数为0.987,说明这两种方法非常一致。
Figure 6. ORF1 assay and serum levels
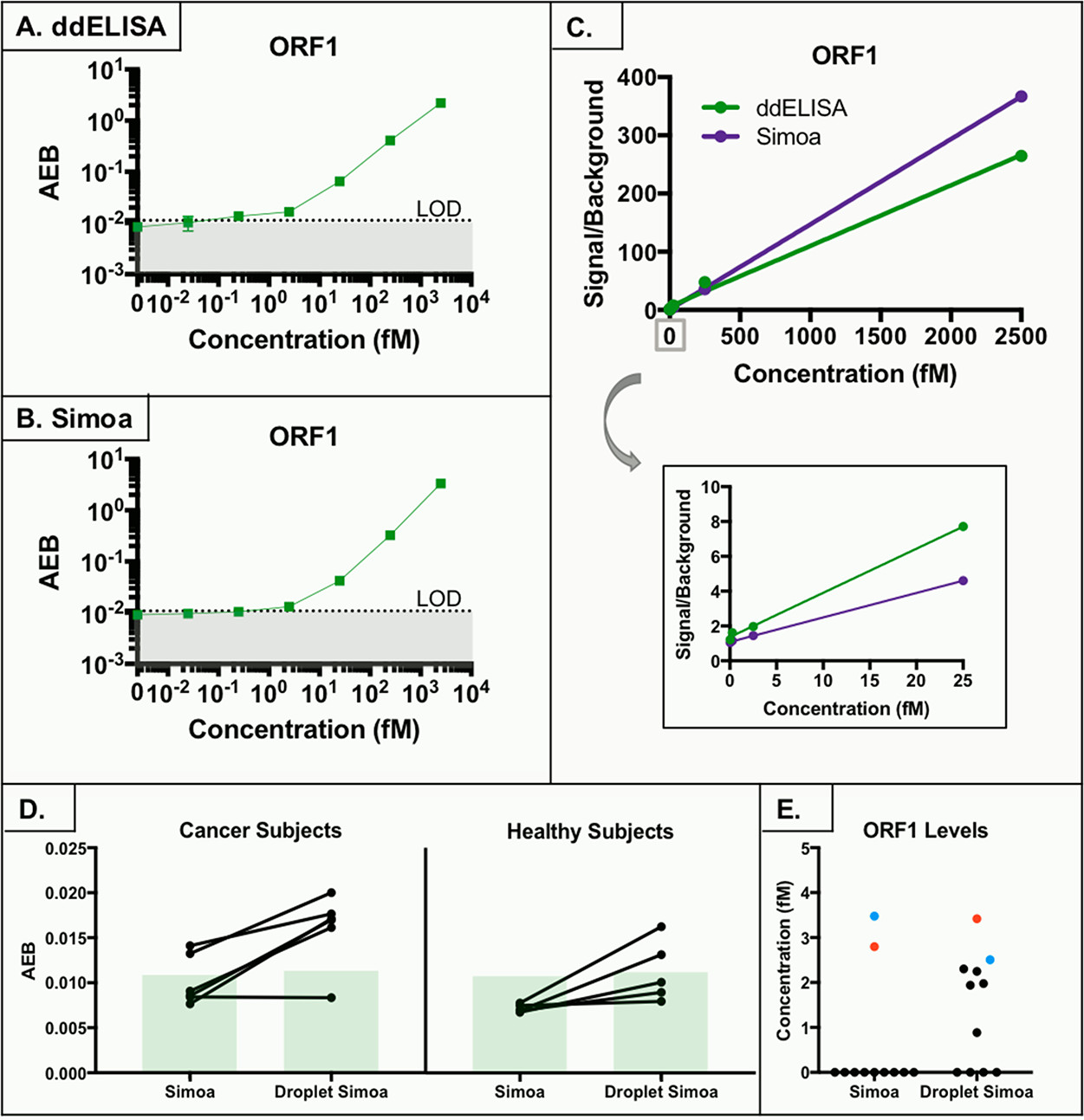
- (A)ORF1的ddELISA校准曲线。
- (B)ORF1的Simoa校准曲线。
- (C)两种方法ORF1相对于背景的信号强度比较,底部为局部放大。可见ddELISA在低浓度下有更好的信号增加,然而高浓度下Simoa表现更好,可能是小珠数目更多。
- (D)用Simoa和ddELISA测定癌症和健康血清中ORF1的AEB值。
- (E)健康样本中,ddELISA测出2项,Simoa测出0项;癌症样本中,ddELISA测出5项,Simoa测出2项。相同样本被标记为一个颜色。
Conclusion
- 减少了对液珠阵列的成像时间,并且可以使用广域荧光成像方法。
- 减少了检测荧光信号的时间。因为ddELISA容量是皮升级,而Simoa是飞升级而使用于酶-底物反应产生荧光产物的时间也相应增加。
Other Information
密度梯度离心
本试验用的试剂为Optiprep。
亦称平衡密度梯度离心法。用超离心机对小分子物质溶液,长时间加一个离心力场达到沉降平衡,在沉降池内从液面到底部出现一定的密度梯度。若在该溶液里加入少量大分子溶液,则溶液内比溶剂密度大的部分就产生大分子沉降,比溶剂密度小的部分就会上浮,最后在离心力和浮力平衡的位置,集聚形成大分子带状物。利用这种现象,测定核酸或蛋白质等的浮游密度,或根据其差别进行分析的一种沉降平衡法。
Reference
Cohen L, Cui N, Cai Y, et al. Single Molecule Protein Detection with Attomolar Sensitivity Using Droplet Digital Enzyme-Linked Immunosorbent Assay[J]. ACS Nano, American Chemical Society, 2020, 14(8): 9491–9501.